An important consideration for any fMRI experimental design is how efficiently the BOLD response will be estimated, given the timing of events and jittering between trials of interest. Therefore, before doing any scanning, it is useful to do a dry run of the experimental task on a computer outside of the scanner, and then feed this timing information into the software analysis package that you will use. For example, in SPM this would involve Specifying the 1st-Level through the SPM GUI, but not estimating it.
In AFNI, this would involve using 3dDeconvolve with the -nodata command, telling AFNI that there is no functional data to read.
#!/bin/tcsh
3dDeconvolve -nodata 205 3 \
-num_stimts 18 \
-stim_times 1 stimuli/reg1.txt 'GAM' \
-stim_label 1 reg1 \
-stim_times 2 stimuli/reg2.txt 'GAM' \
-stim_label 2 reg2 \
-stim_times 3 stimuli/reg3.txt 'GAM' \
-stim_label 3 reg3 \
-stim_times 4 stimuli/reg4.txt 'GAM' \
-stim_label 4 reg4 \
-stim_times 5 stimuli/reg5.txt 'GAM' \
-stim_label 5 reg5 \
-stim_times 6 stimuli/reg6.txt 'GAM' \
-stim_label 6 reg6 \
-stim_times 7 stimuli/reg7.txt 'GAM' \
-stim_label 7 reg7 \
-stim_times 8 stimuli/reg8.txt 'GAM' \
-stim_label 8 reg8 \
-stim_times 9 stimuli/reg9.txt 'GAM' \
-stim_label 9 reg9 \
-stim_times 10 stimuli/reg10.txt 'GAM' \
-stim_label 10 reg10 \
-stim_times 11 stimuli/reg11.txt 'GAM' \
-stim_label 11 reg11 \
-stim_times 12 stimuli/reg12.txt 'GAM' \
-stim_label 12 reg12 \
-stim_times 13 stimuli/reg13.txt 'GAM' \
-stim_label 13 reg13 \
-stim_times 14 stimuli/reg14.txt 'GAM' \
-stim_label 14 reg14 \
-stim_times 15 stimuli/reg15.txt 'GAM' \
-stim_label 15 reg15 \
-stim_times 16 stimuli/reg16.txt 'GAM' \
-stim_label 16 reg16 \
-stim_times 17 stimuli/reg17.txt 'GAM' \
-stim_label 17 reg17 \
-stim_times 18 stimuli/reg18.txt 'GAM' \
-stim_label 18 reg18 \
-x1D X.xmat.1D -xjpeg X.jpg \
-x1D_uncensored X.nocensor.xmat.1D \
(A couple notes about the above code: 3dDeconvolve -nodata should be followed by the number of time points, and the TR used. In this example, I had 205 TRs, with a TR of 3 seconds, for a total of 615 seconds per run. Also, the names of the regressors have been changed to generic labels.)
The output of this command will produce estimates of how efficiently 3dDeconvolve is able to fit a model to the data provided. The Signal+Baseline matrix condition is especially important, as any singularities in this matrix (e.g., perfect or very high correlations between regressors) will make any solution impossible, as this implies that there are potentially infinite answers to estimating the beta weights for each condition. You should see either GOOD or VERY GOOD for each matrix condition; anything less than that will require using the -GOFORIT command, which can override matrix warnings. However, this should be done with caution, and only if you know what you are doing. Which I don't, most of the time.
Also notice that a number labeled the "norm. std. dev." is calculated, which is the root mean square of the measurement error variance for each condition (not to be confused with plain variance). Higher unexplained variance is less desirable, and apparently this design results in very high unexplained variance. More details about how to mitigate this, as well as a more thorough discussion of measurement error variance in general, can be found in this AFNI tutorial.
This command also produces an X.xmat.1D file, which can then be used to either visualize the regressors via 1dplot, or to examine correlations between regressors. This is done with 1d_tool.py, which is the default in the uber_subject.py stream. 1d_tool.py will set the cutoff at 0.4 before it produces any warnings, although this can be changed with the -cormat_cutoff option.
In this example, I compared two designs, one using an exponential distribution of jitters (with more jitters at shorter durations), and another which sampled equally from jitters of different durations.While the default for 1d_tool.py is to show only correlations between regressors at 0.4 or above, this can be changed using the -cormat_cutoff command, as shown below:
This command produced the following output for each design:
Note that while both do not have correlations above 0.4, there are substantial differences when looking at the range of correlations between 0.1 and 0.3. In both cases, however, each design does a relatively good job of reducing correlations between the regressors. This should also be compared against the measurement error variance as described above, as a design may produce higher correlations, but lower measurement error variance.
Final note: There are programs to create efficient designs, such as Doug Greve's optseq and AFNI's RSFgen. However, this will produce designs that are expected to be used for each subject, which potentially could lead to ordering effects. Although the likelihood of this having any significant effect on your results is small, it is still possible. In any case, all of these things should be considered together when designing a scanning study, as it takes far less time to go through these steps than to end up with an inefficient design.
In AFNI, this would involve using 3dDeconvolve with the -nodata command, telling AFNI that there is no functional data to read.
#!/bin/tcsh
3dDeconvolve -nodata 205 3 \
-num_stimts 18 \
-stim_times 1 stimuli/reg1.txt 'GAM' \
-stim_label 1 reg1 \
-stim_times 2 stimuli/reg2.txt 'GAM' \
-stim_label 2 reg2 \
-stim_times 3 stimuli/reg3.txt 'GAM' \
-stim_label 3 reg3 \
-stim_times 4 stimuli/reg4.txt 'GAM' \
-stim_label 4 reg4 \
-stim_times 5 stimuli/reg5.txt 'GAM' \
-stim_label 5 reg5 \
-stim_times 6 stimuli/reg6.txt 'GAM' \
-stim_label 6 reg6 \
-stim_times 7 stimuli/reg7.txt 'GAM' \
-stim_label 7 reg7 \
-stim_times 8 stimuli/reg8.txt 'GAM' \
-stim_label 8 reg8 \
-stim_times 9 stimuli/reg9.txt 'GAM' \
-stim_label 9 reg9 \
-stim_times 10 stimuli/reg10.txt 'GAM' \
-stim_label 10 reg10 \
-stim_times 11 stimuli/reg11.txt 'GAM' \
-stim_label 11 reg11 \
-stim_times 12 stimuli/reg12.txt 'GAM' \
-stim_label 12 reg12 \
-stim_times 13 stimuli/reg13.txt 'GAM' \
-stim_label 13 reg13 \
-stim_times 14 stimuli/reg14.txt 'GAM' \
-stim_label 14 reg14 \
-stim_times 15 stimuli/reg15.txt 'GAM' \
-stim_label 15 reg15 \
-stim_times 16 stimuli/reg16.txt 'GAM' \
-stim_label 16 reg16 \
-stim_times 17 stimuli/reg17.txt 'GAM' \
-stim_label 17 reg17 \
-stim_times 18 stimuli/reg18.txt 'GAM' \
-stim_label 18 reg18 \
-x1D X.xmat.1D -xjpeg X.jpg \
-x1D_uncensored X.nocensor.xmat.1D \
(A couple notes about the above code: 3dDeconvolve -nodata should be followed by the number of time points, and the TR used. In this example, I had 205 TRs, with a TR of 3 seconds, for a total of 615 seconds per run. Also, the names of the regressors have been changed to generic labels.)
The output of this command will produce estimates of how efficiently 3dDeconvolve is able to fit a model to the data provided. The Signal+Baseline matrix condition is especially important, as any singularities in this matrix (e.g., perfect or very high correlations between regressors) will make any solution impossible, as this implies that there are potentially infinite answers to estimating the beta weights for each condition. You should see either GOOD or VERY GOOD for each matrix condition; anything less than that will require using the -GOFORIT command, which can override matrix warnings. However, this should be done with caution, and only if you know what you are doing. Which I don't, most of the time.
Also notice that a number labeled the "norm. std. dev." is calculated, which is the root mean square of the measurement error variance for each condition (not to be confused with plain variance). Higher unexplained variance is less desirable, and apparently this design results in very high unexplained variance. More details about how to mitigate this, as well as a more thorough discussion of measurement error variance in general, can be found in this AFNI tutorial.
This command also produces an X.xmat.1D file, which can then be used to either visualize the regressors via 1dplot, or to examine correlations between regressors. This is done with 1d_tool.py, which is the default in the uber_subject.py stream. 1d_tool.py will set the cutoff at 0.4 before it produces any warnings, although this can be changed with the -cormat_cutoff option.
In this example, I compared two designs, one using an exponential distribution of jitters (with more jitters at shorter durations), and another which sampled equally from jitters of different durations.While the default for 1d_tool.py is to show only correlations between regressors at 0.4 or above, this can be changed using the -cormat_cutoff command, as shown below:
1d_tool.py -cormat_cutoff 0.1 -show_cormat_warnings -infile X.xmat.1D
This command produced the following output for each design:
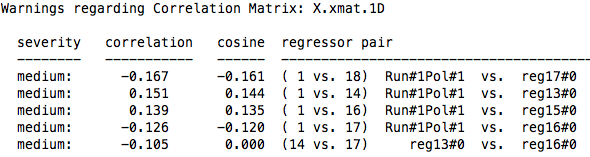 |
| Equal Jitters |
 |
| Exponential Jitters |
Note that while both do not have correlations above 0.4, there are substantial differences when looking at the range of correlations between 0.1 and 0.3. In both cases, however, each design does a relatively good job of reducing correlations between the regressors. This should also be compared against the measurement error variance as described above, as a design may produce higher correlations, but lower measurement error variance.
Final note: There are programs to create efficient designs, such as Doug Greve's optseq and AFNI's RSFgen. However, this will produce designs that are expected to be used for each subject, which potentially could lead to ordering effects. Although the likelihood of this having any significant effect on your results is small, it is still possible. In any case, all of these things should be considered together when designing a scanning study, as it takes far less time to go through these steps than to end up with an inefficient design.

