DTI data comes into this world just as we do - raw, unprocessed, unthresholded, and disgusting. Today's parents are much too easy on their children; back in my day, boys and girls were expected to correct their own erratic motion artifacts, to strip their own skulls when too much cranium started sprouting around their cortex, and reach significance through their own diligence and hard work. "No dinner for you until you've gotten rid of those filthy distortion currents on your hands, young man!" was a common saying around my household.
Unfortunately, these days we live in a society increasingly avoiding taking responsibility for their own actions, filled with people more and more unwilling to strip their own skulls. (Even though that would save neuroimaging researchers countless hours of preprocessing.) This trend is the result of several complex cultural and sociological factors, by which I mean: Lawyers. Things have gotten so bad that you could, to take a random example, potentially sue someone posting advice on an unlicensed website if following those instructions somehow messed up your data.
This actually happened to a friend of mine who publishes the nutrition blog tailored for Caucasian males, White Man's Secret. After one of his readers formed kidney stones as a result of following advice to drink a two-liter of Mello Yello every day, the author was sued for - and I quote from the court petition - "Twelve Bazillion Dollars."
In any case, DTI data takes some time to clean up before you can do any statistics on it. However, the preprocessing steps are relatively few, and are analogous to what is done in a typical FMRI preprocessing pipeline. The steps are to identify and correct any distortions during the acquisition, and then to correct for eddy currents.
The first step, distortion correction, can be done with FSL's topup command. Note that this will require two diffusion weighted images, one with the encoding direction in anterior-to-posterior direction (AP) and one encoded in the posterior-to-anterior direction (PA). You will also need a textfile containing information about the encoding directions and the total readout time (roughly the amount of time between the first echo and the last echo); for example, let's say we have a text file, acqparams.txt, containing the following rows:
0 -1 0 0.0665
0 1 0 0.0665
Here a -1 means AP, and 1 means PA. The last column is the readout time. If you have any questions about this, ask your scanner technician. "What if he's kind of creepy?" I don't care, girl; you gotta find out somehow.
While you're at it, also ask which image from the scan was the AP scan, and which was the PA scan. Once you have identified each, you should combine the two using fslmerge:
fslmerge -t AP_PA_scan AP_scan PA_scan
Once you have that, you have all you need for topup:
topup --imain=AP_PA_scan --datain=acqparams.txt --out=topup_AP_PA_scan
This will produce an image, topup_AP_PA_scan, of where the distortions are located in your DTI scan. This will then be called by the eddy tool to undo any distortions at the same time that it does eddy correction.
The only other preparation you need for the eddy correction is an index list of where the acquisition parameters hold (which should usually just be a 1 for each row for each scan), and a mask excluding any non-brain content, which you can do with bet:
bet dwi_image betted_dwi_image -m -f 0.2
Then feed this into the eddy command (note that a bvecs and bvals image should be automatically generated if you imported your data using a tool such as dcm2niigui):
eddy --imain=dwi_image --mask=betted_dwi_image --index=index.txt --acqp=acqparams.txt --bvecs=bvecs --bvals=bvals --fwhm=0 --topup=topup_AP_PA_scan --flm=quadratic --out=eddy_unwarped_images
Note that the --fwhm and --flm arguments I chose here are the defaults that FSL recommends.
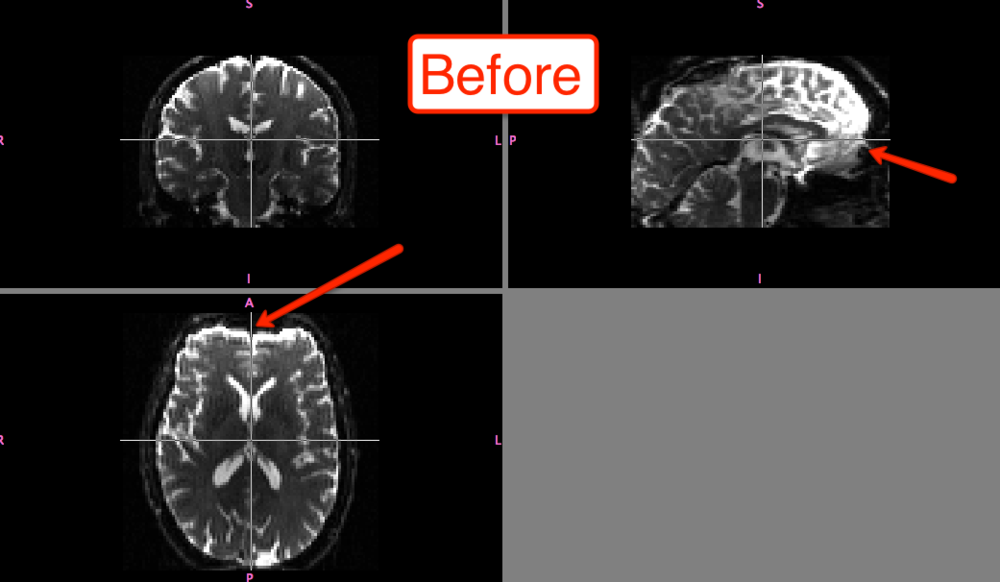
The differences before and after can be seen in fslview:
Of course, the distortion correction step isn't strictly necessary; and you can still do a much simpler version of eddy correction by simply selecting FDT Diffusion from the FSL menu and then selecting the Eddy Correction tab, which only requires input and output arguments. All I'm saying is that, if you're feeling particularly litigious, you should probably be safe and do both steps.