A couple of weeks ago, alert reader Paula DiNoto asked about how to output SPM files in NIFTI format instead of ANALYZE (.hdr/.img). Apparently, for those of you bleeding-edge adrenaline junkies who just have to have the latest thing, SPM12 doesn't give you this option in the interface. SPM5, however - you know, the one that the rest of us barbarians use - still does. (I'll save my gloating for when I meet you face-to-face at the conferences.)
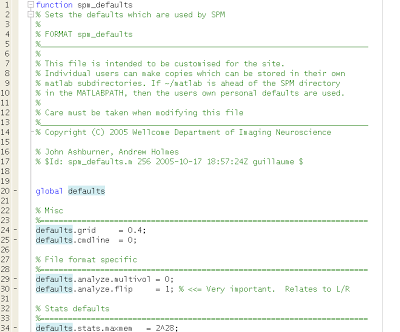
Getting back to the story, I had to admit to Paula that I was, for the first time in my life, unable to figure out how to troubleshoot the problem. However, this was merely a ruse to get Paula motivated; she ended up solving the problem by changing "spm_defaults.m" and changing the default.images.format field to 'nii'. Obviously I already knew this, but I was pleased to see her figure it out for herself.
Thanks again to Paula, whom I once called, in a moment of self-forgetfulness, the most wonderful person I had ever met.