Power is like the sun: Everybody wants it, everybody finds in it a pleasant burning sensation, and yet nobody really knows what it is or how to get more of it. In my introductory statistics courses, this was the one concept - in addition to a few other small things, like standard error, effect size, sampling distributions, t-tests, and ANOVAs - that I completely failed to comprehend. Back then, I spoke as a child, I understood like a child, I reasoned like a child; but then I grew older, and I put away childish things, and resolved to learn once and for all what power really was.
1. What is Power?
The textbook definition of statistical power is rejecting the null hypothesis when it is, in fact, false; and everyone has a vague sense that, as the number of subjects increases, power increases as well. But why is this so?
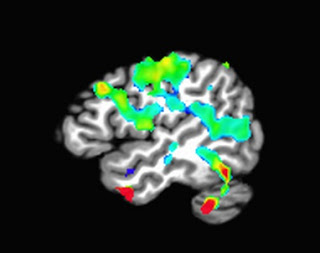
To illustrate the concept of power, consider two partially overlapping distributions, shown in blue and red:
The blue distribution is the null distribution, stating that there is no effect, or difference; the red distribution, on the other hand, represents the alternative hypothesis that there is some effect or difference. The red dashed line represents our rejection region, beyond which we would reject the null hypothesis; and we can see that the more density of the alternative distribution that lies outside of this cutoff region, the greater probability we have of randomly drawing a sample that leads to a rejection of the null hypothesis, and therefore the greater our statistical power.
However, the sticking point is this: How do we determine where to place our alternative distribution? Potentially, it could be anywhere. So how do we decide where to put it?
One approach is to make an educated guess; and there is nothing wrong with this approach, given that it is solidly based on theory, and this may be appropriate if you do not have the time or resources to run an adequate pilot sample to do a power calculation. Another approach may be to estimate the mean of the alternative distribution based on the results from other studies; but, assuming that those results were significant, they have a greater probability of being sampled from the upper tail of the alternative distribution, and therefore have a larger probability of being greater than the true mean of the alternative distribution.
A third approach is to estimate the mean of the alternative distribution based on a sample - which is the logic behind doing a pilot study. This is often the best estimate we can make of the alternative distribution, and, given that you have the time and resources to carry out such a pilot study, is the best option for estimating power.
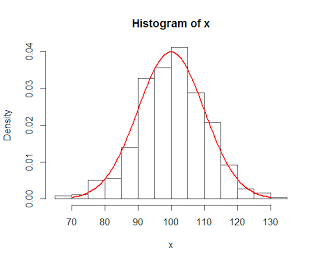
Once the mean of the alternative distribution has been established, the next step is to determine how power can be affected by changing the sample size. Recall that the standard error, or standard deviation of your sampling distribution of means, is inversely related to the square root of the number of subjects in your sample; and, critically, that the standard error is assumed to be the same for
both the null distribution and the alternative distribution. Thus, increasing the sample size leads to a reduction in the spread of both distributions, which in turn leads to less overlap between the two distributions and again increases power.
 |
| Result of increasing the sample size from 4 to 10. Note that there is now less overlap between the distributions, and that more of the alternative distribution now lies to the right of the cutoff threshold, increasing power. |
2. Power applied to FMRI
This becomes an even trickier issue when dealing with neuroimaging data, when gathering a large number of pilot subjects can be prohibitively expensive, and the funding of grants depends on reasonable estimates from a power analysis.
Fortunately, a tool called
fmripower allows the researcher to calculate power estimates for a range of potential future subjects, given a small pilot sample. The interface is clean, straightforward, and easy to use, and the results are useful not only for grant purposes, but also for a sanity check of whether your effect will have enough power to warrant going through with a full research study. If achieving power of about 80% requires seventy or eighty subjects, you may want to rethink your experiment, and possibly collect another pilot sample that includes more trials of interest or a more powerful design.
A few caveats about using fmripower:
- This tool should not be used for post-hoc power analyses; that is, calculating the power associated with a sample or full dataset that you already collected. This type of analysis is uninformative (since we cannot say with any certainty whether our result came from the null distribution or a specific alternative distribution), and can be misleading (see Hoenig & Heisey, 2001).
- fmripower uses a default atlas when calculating power estimates, which parcellates cortical and subcortical regions into dozens of smaller regions of interest (ROIs). While this is useful for visualization and learning purposes, it is not recommended to use every single ROI; unless, of course, you correct for the number of ROIs used by applying a method such as Bonferroni correction (e.g., dividing your Type I error rate by the number of ROIs used).
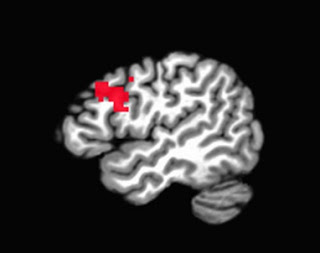
- When selecting an ROI, make sure that it is independent (cf. Kriegeskorte, 2009). This means choosing an ROI based on either anatomical landmarks or atlases, or from an independent contrast (i.e., a contrast that does not share any variance or correlate with your contrast of interest). Basing your ROI on your pilot study's contrast of interest - that is, the same contrast that you will examine in your full study - will bias your power estimate, since any effect leading to significant activation in a small pilot sample will necessarily be very large.
- For your final study, do not include your pilot sample, as this can lead to an inflated Type I error rate (Mumford, 2012). A sample should be used for power estimates only; it should not be included in the final analysis.
Once you've experimented around with fmripower and gotten used to its interface, either with SPM or FSL, simply load up your group-level analysis (either FSL's cope.feat directory and corresponding cope.nii.gz file for the contrast of interest, or SPM's .mat file containing the contrasts in that second-level analysis), choose an unbiased ROI, your Type I error rate, and whether you want to estimate power for a specific subject number or across a range of subjects. I find the range much more useful, as it gives you an idea of the sweet spot between the number of subjects and power estimate.
Thanks to Jeanette Mumford, whose obsession with power is an inspiration to us all.