As part of a new course in neuroimaging methods at Haskins Laboratories, I've begun updating videos on topics such as functional connectivity, context-dependent correlations, and how to accept bribes as a reviewer. The first topic we've covered is resting-state functional connectivity, a sophisticated-sounding name designed to make the subject think he is doing something of immense scientific importance by lying still and doing nothing, when in reality it's to distract him while we find out how to hell to hook up the experimental laptop.
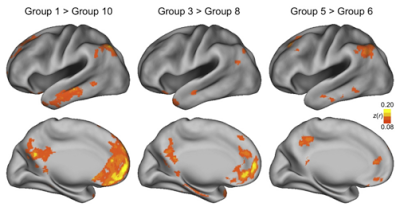
Aside from its usefulness as a stall tactic, resting-state connectivity can also reveal resting-state networks, or correlations between the signal of distant regions of the brain. This provides clues to how structural connectivity - i.e., white matter connections - interact with the BOLD signal, as well as whether differences in resting-state connectivity is a marker for mental disorders such as Alzheimer's or schizophrenia.
The following video takes you step-by-step through functional connectivity analysis, using an online dataset from openfmri.org. One major change from my previous tutorials is condensing all the information into one long video, and providing time markers for each segment in the "Show More" box. This way the viewer can jump around to the information that they need, without having to keep track of several different videos detailing different steps. I hope it's an improvement, and I would like to get feedback.
I've also posted the lecture on resting-state analysis given at Haskins Laboratories on November 3rd. You won't learn much new here that isn't in the video above, but it does have more information. For most of the lecture you can only see the top of my head bobbing around, but that's OK. Eyes on the slides, not the hair.
Exercises:
Aside from its usefulness as a stall tactic, resting-state connectivity can also reveal resting-state networks, or correlations between the signal of distant regions of the brain. This provides clues to how structural connectivity - i.e., white matter connections - interact with the BOLD signal, as well as whether differences in resting-state connectivity is a marker for mental disorders such as Alzheimer's or schizophrenia.
The following video takes you step-by-step through functional connectivity analysis, using an online dataset from openfmri.org. One major change from my previous tutorials is condensing all the information into one long video, and providing time markers for each segment in the "Show More" box. This way the viewer can jump around to the information that they need, without having to keep track of several different videos detailing different steps. I hope it's an improvement, and I would like to get feedback.
I've also posted the lecture on resting-state analysis given at Haskins Laboratories on November 3rd. You won't learn much new here that isn't in the video above, but it does have more information. For most of the lecture you can only see the top of my head bobbing around, but that's OK. Eyes on the slides, not the hair.
Exercises:
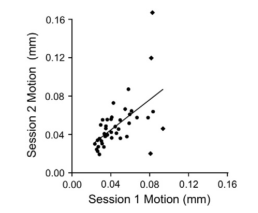
- Set the errts dataset as the underlay, and select "Graph". From the "Opt" menu, select "Write Center." Rename the output 1D file, and use 1dplot to see the timecourse. This can be used as a seed for another connectivity analysis.
- Other resting state networks include the somatosensory network, the visual network, and the language network. Research one of these networks, determine where the hubs are, and run a resting state analysis on a seed placed in that hub.
- Run correlations for a group of subjects, convert to z-scores, and do a second-level t-test using uber_ttest.py.
- Modify the afni_proc.py script to apply 3dRSFC to your data (see Example 10b in afni_proc.py -help)